Molecular Transducers of Human Skeletal Muscle Remodeling under Different Loading States
Tanner Stokes cell reports. 2020.
Highlights
• Muscle loading and unloading regulate the untranslated regions of mRNA
• Regulated genes form functional networks central to muscle plasticity
• 141 genes correlate with muscle growth in 3 independent validation cohorts
• Several growth-correlated network genes directly regulate myocyte protein synthesis
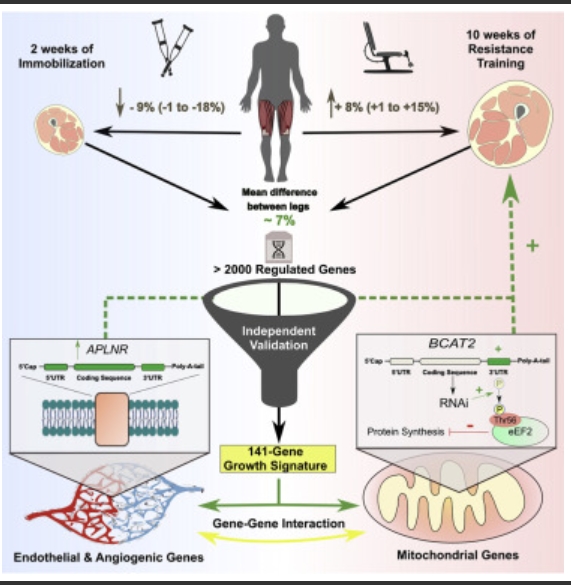
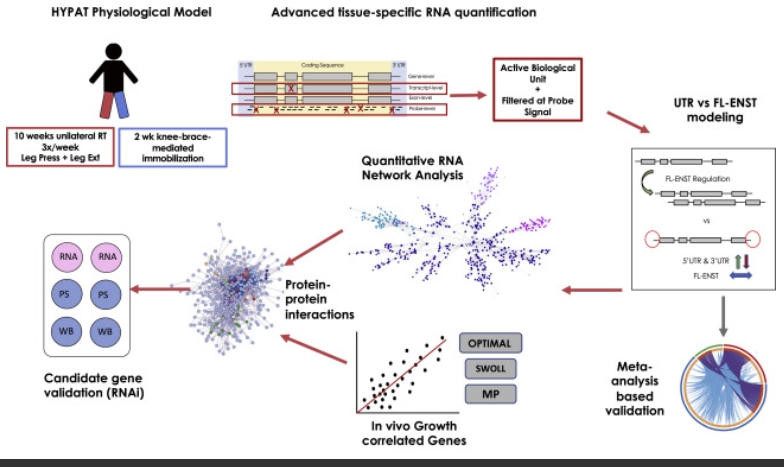
Loading of skeletal muscle changes the tissue phenotype reflecting altered metabolic and functional demands. In humans, heterogeneous adaptation to loading complicates the identification of the underpinning molecular regulators. A within-person differential loading and analysis strategy reduces heterogeneity for changes in muscle mass by ∼40% and uses a genome-wide transcriptome method that models each mRNA from coding exons and 3′ and 5′ untranslated regions (UTRs). Our strategy detects ∼3–4 times more regulated genes than similarly sized studies, including substantial UTR-selective regulation undetected by other methods.
We discover a core of 141 genes correlated to muscle growth, which we validate from newly analyzed independent samples (n = 100). Further validating these identified genes via RNAi in primary muscle cells, we demonstrate that members of the core genes were regulators of protein synthesis. Using proteome-constrained networks and pathway analysis reveals notable relationships with the molecular characteristics of human muscle aging and insulin sensitivity, as well as potential drug therapies.